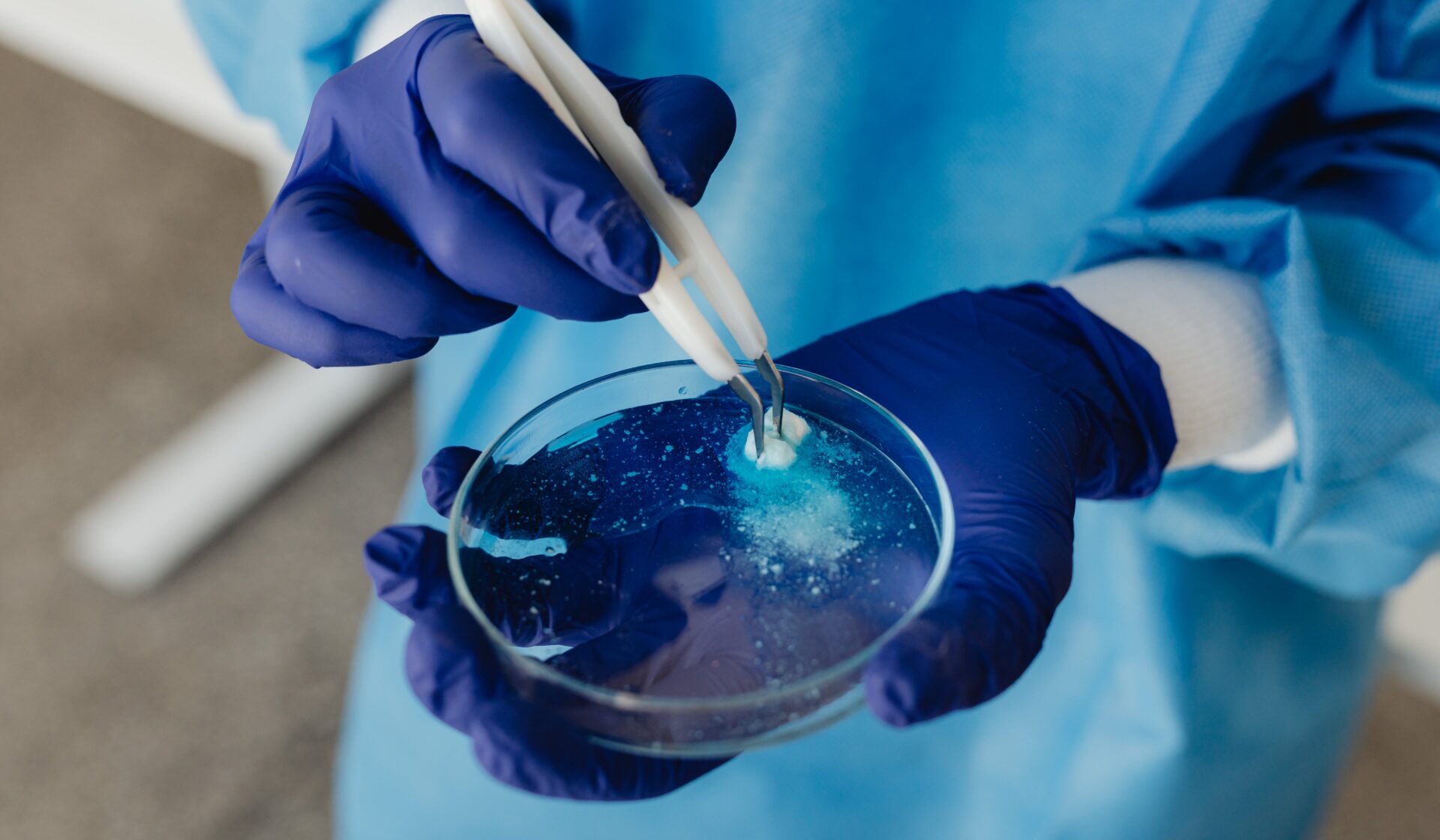
A deep-learning algorithm has helped scientists identify new compounds that are effective against antibiotic-resistant bacteria – a public health threat that causes thousands of deaths annually.
With doctors concerned that antibiotics have become increasingly ineffective since Fleming first purified penicillin in 1928, the recent discovery of a compound which can kill the drug-resistant bacteria that’s responsible for thousands of deaths worldwide every year is a welcome one.
Using a deep-learning algorithm, scientists could identify the first new antibiotic in over half a century, demonstrating the potential of artificial intelligence in the medical field and posing a solution to antibiotic resistance, which is among the biggest global threats to human health.
To refine the selection of this new class of antibiotic candidates, a team at the laboratory of James Collins of the Broad Institute of the Massachusetts Institute of Technology and Harvard University used a type of AI known as deep-learning to screen 12 million compounds for antibiotic activity.
After analysing them through computer simulations, the trained AI models then found 3646 compounds with ideal drug-like properties.
.@delafuenteupenn shared how artificial intelligence (#AI) can help guide the development of new #antibiotics, including new findings that use the concept of deep learning to discover new antibiotics, a practice employed by de la Fuente’s lab. @sciam https://t.co/eFGmY0ZLHo
— Penn Medicine (@PennMedicine) December 26, 2023
Additional calculations identified the chemical substructures that could explain each compound’s properties (namely whether or not they’re harmful to the human body), which scientists compared before testing 238 of them on mice.
In doing so, they uncovered five different non-toxic ones that showed significant promise against methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus – which are some of the most stubbornly hard-to-kill pathogens that we know of.
‘What we set out to do in this study was to open the black box,’ says Felix Wong, an author of the study which was published in Nature last month.
‘Our [AI] models tell us not only which compounds have selective antibiotic activity, but also why, in terms of their chemical structure.’
The development builds on previous research into the power of technology to combat the ongoing antibiotic-resistance crisis.
In the case of deep-learning, scientists have been using it more and more to accelerate the identification of potential drug candidates, predict their properties, and optimise the process of getting them to patients in need.
‘These models consist of very large numbers of calculations that mimic neural connections, and no one really knows what’s going on underneath the hood,’ continues Wong.
‘Our work provides a framework that is time-efficient, resource-efficient, and mechanistically insightful, from a chemical-structure standpoint, in ways that we haven’t had to date.’